Articles
- Page Path
- HOME > Epidemiol Health > Volume 42; 2020 > Article
-
Original Article
Copro-molecular diagnosis of the Toxoplasmatinae subfamily in dog and cat populations in northern Iran -
Leila Izadi1,2,3
 , Shahabeddin Sarvi1,2
, Shahabeddin Sarvi1,2 , Seyed Abdollah Hosseini1,2
, Seyed Abdollah Hosseini1,2 , Afsaneh Amouei1,2
, Afsaneh Amouei1,2 , Mehdi Sharif1,4
, Mehdi Sharif1,4 , Mohammad Taghi Rahimi5
, Mohammad Taghi Rahimi5 , Tooran Nayeri1,2,3
, Tooran Nayeri1,2,3 , Ahmad Daryani1,2
, Ahmad Daryani1,2
-
Epidemiol Health 2020;42:e2020074.
DOI: https://doi.org/10.4178/epih.e2020074
Published online: December 4, 2020
1Toxoplasmosis Research Center, Mazandaran University of Medical Sciences, Sari, Iran
2Department of Parasitology and Mycology, School of Medicine, Mazandaran University of Medical Science, Sari, Iran
3Student Research Committee, Mazandaran University of Medical Science, Sari, Iran
4Department of Parasitology, School of Medicine, Sari Branch, Islamic Azad University, Sari, Iran
5School of Medicine, Shahroud University of Medical Sciences, Shahroud, Iran
- Correspondence: Ahmad Daryani Department of Medical Parasitology, School of Medicine, Mazandaran University of Medical Sciences, PC 48168-95475, Sari, Iran E-mail: daryanii@yahoo.com
©2020, Korean Society of Epidemiology
This is an open-access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
OBJECTIVES
- The oocysts of the Toxoplasmatinae subfamily (Neospora caninum, Hammondia hammondi and H. heydorni, and Besnoitia besnoiti) are morphologically similar to Toxoplasma gondii, and indistinguishable from each other. This study investigated the prevalence of the Toxoplasmatinae subfamily in dog and cat fecal samples using a nested polymerase chain reaction method.
-
METHODS
- Overall, 200 fecal samples from domestic dogs (n=120) and cats (n=80) were collected from 15 farms in northern Iran. The samples were homogenized in 2.5% potassium dichromate solution and subsequently concentrated with sucrose solution. DNA was extracted from samples using a genomic DNA kit. Specific primers and the 18S rDNA gene were used to screen and detect all Toxoplasmatinae oocysts.
-
RESULTS
- Overall, 2.5% (3 of 120) and 22.5% (18 of 80) of the fecal samples collected from dogs and cats were infected with Toxoplasmatinae. In dogs, 2 samples were positive for N. caninum and 1 sample was positive for T. gondii. In cats, all 18 positive samples belonged to T. gondii. No contamination with H. heydorni was observed in dog fecal samples or H. hammondi and B. besnoiti in cat fecal samples. A phylogenetic analysis revealed that the T. gondii (cat) and N. caninum (dog) found had similarities with parasites reported from other regions of the world.
-
CONCLUSIONS
- This is the first study to provide data on the epidemiology of Toxoplasmatinae oocysts in Iran. The findings suggest that public-health monitoring for the effective control of feces from cats and dogs and improved pet hygiene habits are needed.
- Tissue-cyst-forming apicomplexan parasites, such as Toxoplasma gondii, Neospora caninum, Hammondia spp. (H. hammondi and H. heydorni), and Besnoitia besnoiti belong to the Toxoplasmatinae subfamily, which is associated with a variety of diseases in humans and other animals [1].
- T. gondii is an obligate intracellular protozoan that causes the infectious disease toxoplasmosis. It has 2 distinct life cycles. The sexual cycle occurs only in cats, the definitive host. The asexual cycle occurs in all warm-blooded animals (mammals and birds) and humans. This parasite is pathogenic in human and animals [2]. It is often asymptomatic, but it can lead to severe complications in infants and persons with acquired immune deficiency, especially acquired immune deficiency syndrome (AIDS); its symptoms can include microcephaly, hydrocephalus, and chorioretinitis in infants, as well as repeated attacks of encephalitis in immunodeficient individuals, which may even be fatal. Clinical toxoplasmosis in animals is seen most often in sheep and goats when infected during pregnancy and manifests as abortions, stillbirths, and mummification [3].
- N. caninum is an obligate intracellular coccidian parasite that causes spontaneous abortion in infected livestock and paralysis in dogs [4,5]. Dogs are currently believed to be the definitive host of N. caninum. Intermediate hosts such as cattle can be infected by neosporosis via ingestion of oocysts from the feces of dogs (horizontal transmission) or from the dam to the fetus (vertical transmission). Despite serological evidence of human exposure in immunocompromised individuals, this unicellular organism is not considered to be a zoonotic parasite [6,7].
- H. hammondi and H. heydorni are cyst-forming coccidian protozoans, with very low prevalence rates. Cats are the definitive hosts of H. hammondi and dogs, foxes, and coyotes are considered to be the definitive hosts of H. heydorni. Unlike T. gondii and N. caninum, H. hammondi and H. heydorni are not known to lead to clinical signs in either their definitive or intermediate hosts [8].
- B. besnoiti is the etiological agent of bovine besnoitiosis and has a significant economic impact on the cattle industry. Currently, knowledge of the definitive host for B. besnoiti is limited. However, it seems that cats are the definitive hosts for several Besnoitia species that infect wildlife, and cattle represent an important intermediate host for this protozoan [9].
- All oocysts of the Toxoplasmatinae subfamily are similar in size (9-14 μm) and morphologically indistinguishable from each other [10,11]. Since their oocysts are physically and morphologically similar to T. gondii, and cannot be distinguished from each other [12], a diagnosis based on light microscopy of oocysts in feces is not a method of choice for species identification of these important coccidia. Instead, these oocysts can only be distinguished via molecular methods. Diagnostic molecular techniques, such as polymerase chain reaction (PCR), are a highly sensitive and specific alternative to morphological methods [12-14]. Given the importance of molecular methods, identification of these parasites is crucial for precise control of these zoonotic diseases and making proper decisions regarding economic losses. The aim of this study was to investigate the infection rates of the Toxoplasmatinae subfamily in fecal samples of dogs and cats as reservoir hosts, using nested PCR followed by sequencing and a phylogenetic assay, to demonstrate the circulation of these parasites in northern Iran.
INTRODUCTION
- Sample collection
- Overall, 200 fecal samples (about 20 g) from domestic dogs (n=120) and cats (n=80) were collected between April 2015 and March 2016 from 15 farms (cattle, sheep, goat) in different region of northern Iran. The fecal samples were transferred to the parasitology laboratory of Mazandaran University of Medical Sciences.
- Purification and sporulation of oocysts
- To purify the oocysts, the fecal material was sieved via a strainer (60 meshes) using tap water. Then, for sporulation, fecal samples were mixed with 2.5% potassium dichromate (K2Cr2O7) and incubated at room temperature for 2 weeks.
- Concentration of oocysts
- To concentrate the oocysts, they were first washed 3 times with tap water by centrifugation (300× g) for 3 minutes to remove the K2Cr2O7. Then, 1 g of the fecal sample was transferred into a 10-mL tube mixed with 9 mL of 1 M sucrose. The samples were centrifuged at 800× g for 5 minutes. The middle layer was removed and placed in a fresh micro-tube, and after centrifuging at 800×g for 8 minutes, the supernatant was removed and the resulting pellet washed twice with distilled water by centrifuging at 800× g for 5 minutes. The supernatant was decanted and the pellet was resuspended in distilled water at the final volume and stored at -20°C before processing.
- DNA extraction and molecular assay
- DNA was extracted from all of the dog and cat fecal samples using the a genomic DNA kit (Dena Zist, Mashhad, Iran) according to the manufacturer’s instructions. To screen and identify the Toxoplasmatinae subfamily, nested PCR was performed to amplify fragments of the 18S rDNA gene with external and internal primers. The external primers were Tg18s48R (5′-CCATGCATGTCTAAGTATAAGC-3′) and Tg18s359R (5′-GTTACCCGTCACTGCCAC-3′), which amplified a 312-bp fragment. The internal primers were Tg18s58F (5′-CTAAGTATAAGCTTTTATACGGC-3′) and Tg18s348R (5′-TGCCACGGTAGTCCAATAC-3′), which amplify a 291-bp region of the 18S rDNA gene [15]. Each amplification reaction was set for a total volume of 25 μL, containing 12.5 μL of 1× PCR mix (Taq PCR Master Mix Kit, Ampliqon, Odense, Denmark), 0.1 mM of each of the external and internal primers and 1.0 μL of template DNA. The final volume (25 µL) was reached by adding nuclease-free water. The PCR conditions in both stages were as follows: initial denaturation for 5 minutes at 94°C; 30 cycles of denaturing for 30 seconds at 94°C, annealing for 30 seconds at 60°C, extension for 30 seconds at 72°C; and final extension for 7 minutes at 72°C.
- In the next step, samples that were positive for Toxoplasmatinae subfamily DNA were subsequently subjected to conventional PCR using specific primers for each parasite. For dog feces samples, specific primers for T. gondii, N. caninum, and H. heydorni were used. For cat feces samples, specific primers for T. gondii, N. caninum, H. hammondi, and B. besnoiti were used. Each PCR reaction was carried out in a volume of 25 µL, containing 12.5 μL of 1× PCR mix (Taq PCR Master Mix Kit, Ampliqon), 0.1 mM of each of the specific primers, and 70 ng of template DNA. PCR amplification was performed with a primary denaturing step at 94°C for 5 minutes, 30 cycles of denaturing at 94°C for 30 seconds, a specific annealing temperature for each parasite, and extension at 72°C for 90 seconds. After this process, final extension was performed at 72°C for 10 minutes (Table 1). The PCR products were confirmed by visualization on a 1.5% agarose gel with an ultraviolet transilluminator after staining with SYBR Safe (which dyed the PC products green).
- Sequencing and phylogenetic analysis
- The preparing of sequences was performed using Chromas version 2.33 (http://www.technelysium.com.au/chromas.html). The Multalin program was applied to compare and align the nucleotide sequences with each other. Phylogenetic analysis was performed by constructing a phylogenetic tree using the Neighbor-Joining method in MEGA version 4.0 (https://www.megasoftware.net/mega4/). The bootstrapping method with 1,000 replicates was used to assess the reliability of the phylogenetic tree.
- Ethics statement
- The research presented in this article was approved (No. 1716) by the Deputy of Research, Mazandaran University of Medical Sciences, Sari, Iran.
MATERIALS AND METHODS
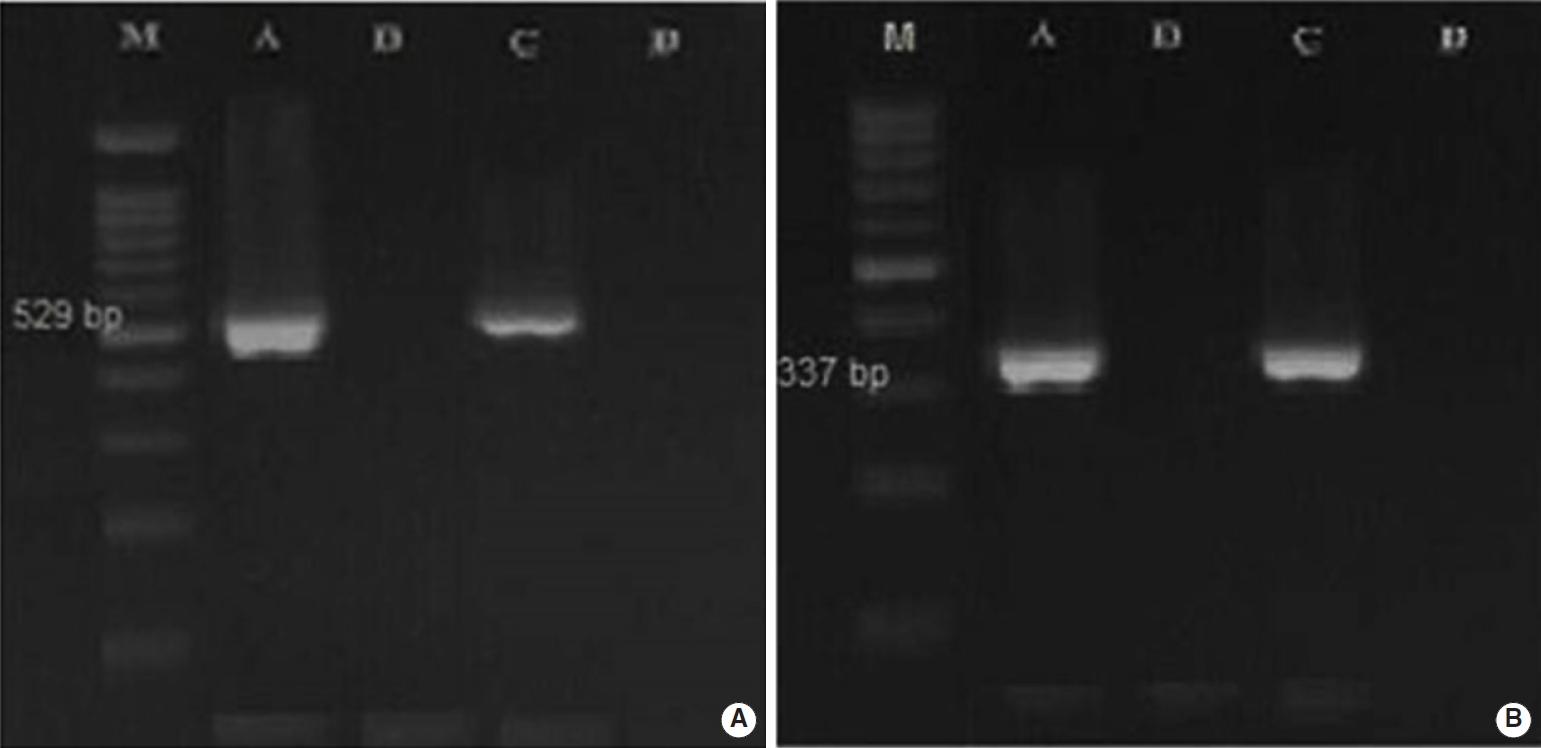
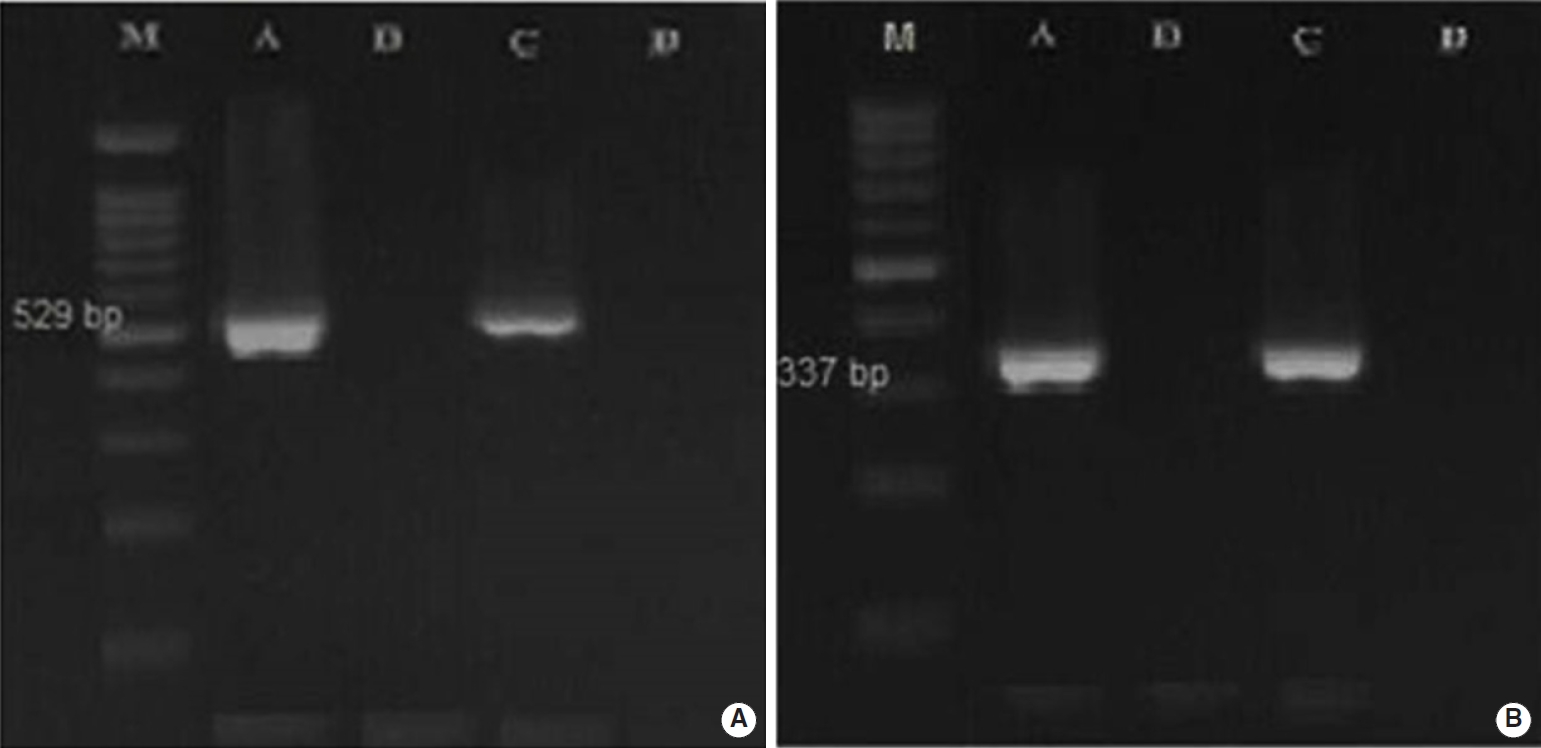
- Overall, 2.5% (3 of 120) and 22.5% (18 of 80) of the fecal samples collected from dogs and cats living on farms in northern Iran were infected with Toxoplasmatinae DNA, respectively, based on nested PCR using the 18S rDNA gene. In the dog fecal samples, the presence of N. caninum and T. gondii DNA was confirmed by the observation a 337-bp band in 2 samples (1.7%) and a 529-bp band in 1 sample (0.8%). However, no contamination with H. heydorni was observed. In cat fecal samples, the presence of T. gondii DNA was confirmed by observation of a 529-bp band in 18 samples (22.5%) (Figure 1). However, no contamination with H. hammondi or B. besnoiti was observed (Table 2).
- Thirteen of the 21 samples of positive PCR products were sequenced by Sanger sequencing, and it was found that 2 samples of N. caninum from the dog fecal samples were registered in the National Center for Biotechnology Information gene bank with accession numbers MN658869 and MN658870. The similarity of T. gondii isolated from the dog fecal samples (MN595275) in comparison with the available genes in the gene data bank was 99.2-99.6%. In addition, 10 sequences of T. gondii isolated from cat fecal samples were deposited in GenBank under the accession numbers MN595276-MN595285.
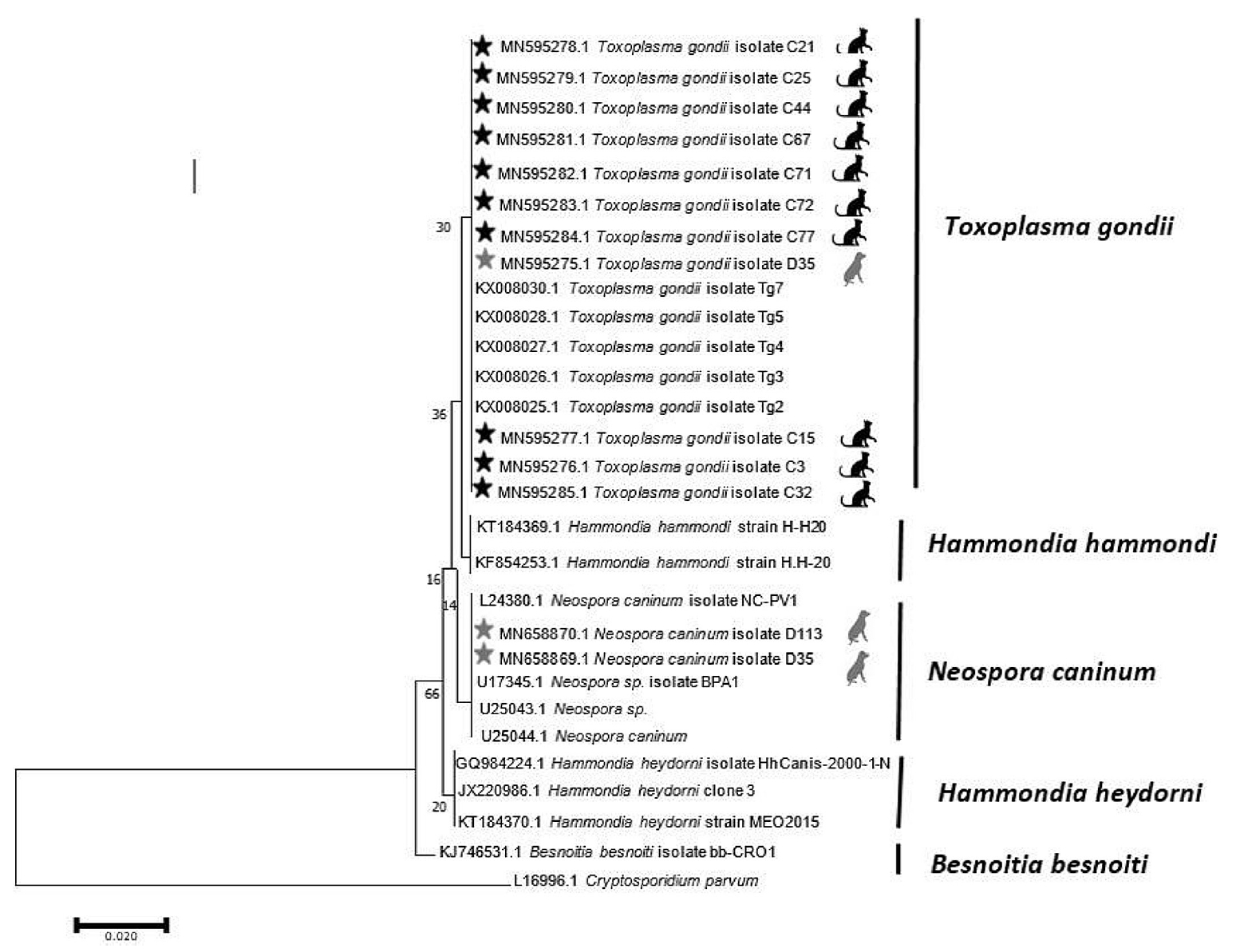
- The findings of the PCR assay closely conformed to those of the Basic Local Alignment Search Tool results and phylogenetic tree. An analysis of the phylogenetic tree revealed that the isolates of T. gondii found in the dog and cat fecal samples in Mazandaran Province were similar to T. gondii isolates from China (accession number: KX008030; host: sheep), Brazil (accession number: KX008028; host: cat), Canada (accession number: KX008027; host: cougar), and France (accession number: KX008026; host: Homo sapiens). Moreover, the N. caninum observed in dog stool was similar to N. caninum isolates reported from the USA (accession number: U17345.1; host: cattle and accession number: U25044.1; host: Canis familiaris) (Figure 2).
RESULTS
- Due to the similarity of Toxoplasmatinae oocysts, microscopic methods cannot distinguish between their oocysts. To solve this problem, molecular methods are recommended. However, T. gondii and N. caninum seropositivity in dogs only indicates contact with the protozoan and does not automatically demonstrate oocyst shedding, PCR has been recommended in recent years for better detection of T. gondii and N. caninum in dogs [20-22].
- PCR is a sensitive and specific assay that has been used to identify neosporosis in both definitive and intermediate hosts [13]. Molecular diagnostic techniques such as PCR are the best choice for detecting H. heydorni from an epidemiological standpoint, and are important for distinguishing this species from other tissue-cyst-forming apicomplexan parasites [23].
- Recent molecular studies based on internal transcribed spacer 1 (ITS1) sequences demonstrated that this gene marker can be used to identify the DNA of apicomplexans that are closely related to T. gondii [13]. It has been shown that N. caninum genomic DNA could be distinguished from H. heydorni using the Np6/N21 primer set. The results of PCR assays using these primers demonstrated that H. heydorni DNA was not amplified, and did not interfere with the amplification of N. caninum DNA in mixed samples. Slapeta and collegues [13,24] and Dubey et al. [5] designed JS4-JS5 primers that are able to specifically amplify the 3’ end of the small subunit rRNA gene and ITS1 of H. heydorni. In this study, we used ITS1 of 18S rDNA as a marker to detect the DNA of apicomplexans that are closely related to T. gondii in cat and dog fecal samples. Our results from dog fecal samples showed that 2.5% (3 of 120) of fecal samples were infected with Toxoplasmatinae DNA. Subsequently, we used the special primers (SJ4/SJ5 and N21+/N6+) to distinguish N. caninum from H. heydorni.
- Our molecular survey using N21+/N6+ primers showed that 1.7% of dog feces samples were infected with N. caninum. The result is similar to that of a study carried out in Australia by King et al. [18], which used molecular methods and found oocysts of N. caninum in 2 out of 132 (1.5%) dog fecal samples. In Iran, 2 studies have been conducted on N. caninum oocysts in dog fecal samples. Razmi [25] in the city of Mashhad, reported a prevalence of N. caninum of 1.5% (2 of 174 samples) using PCR. In another study carried out in western Iran by Ghafarifar et al. [26], 9 positive samples of N. caninum were detected from 428 fecal specimens (2.1%). The prevalence of N. caninum seems to be affected by favorable geographical conditions, the density of the dog population, and the presence of intermediate hosts in the surrounding environment. Dogs can shed oocysts in the environment of farms. N. caninum is considered to be one of the most important infectious agents responsible for abortion in cattle. Exposure of cattle to N. caninum oocysts causes economic losses due to reproductive failure associated with abortion. Therefore, farm owners should be aware of the potential risks posed by the presence of stray dogs around the farm.
- In the present study, H. heydorni was not detected in the fecal samples of domestic dogs. Our results were not unexpected because to our knowledge, the prevalence rate of H. heydorni in dogs in various countries is very low: 0.05% in Germany [4], 0.2% in the Czech Republic [13], and 0.8% in China [27]. However, Li et al. [28] in a rural region of China, reported a seemingly unusually high prevalence of N. caninum in dog feces (17.5%). In our study, T. gondii DNA was surprisingly observed in a feces sample from 1 dog. This result shows that dog feces could pose a risk of toxoplasmosis to other species, including humans; and dogs may serve as mechanical vectors for this parasite [29]. Since dogs are not the definitive host for T. gondii, and its sexual cycle has not been observed, they are not able to shed oocysts. Therefore, in the context of this cross-sectional study, it seems that the dog had ingested T. gondii oocysts by coprophagia [4]. Our results of cat fecal samples showed that 22.5% (18/80) of the samples were infected with Toxoplasmatinae DNA. We subsequently used special primers (TOX4/TOX5, Hham3F/Hham3R, ITS-1F/ITS-1R) to distinguish T. gondii from H. hammondi and B. besnoiti. In the cat fecal samples, the presence of T. gondii DNA was confirmed by the observation of a 529-bp band in 18 samples (22.5%), while no contamination with H. hammondi or B. besnoiti was observed.
- The prevalence of T. gondii in the present study in cats is very similar to that reported by Mancianti et al. [30], who reported a prevalence of 16% (8 positive out of 50 cat fecal samples) in a rural area of Italy. Overall, the prevalence of T. gondii in cat fecal samples in most studies ranges from 0% to 5%. In a study by Berger-Schoch et al. [31] in Switzerland, the prevalence of this parasite was found to be 0.4% using molecular PCR. Jokelainen et al. [32] also reported a prevalence of 1.5% in Finland using microscopic and molecular methods. According to a literature review, the highest prevalence of this parasite in cat fecal samples was reported by Dubey et al. [33] in Ethiopia, with a prevalence of 19.4%. According to the above findings, the observed prevalence of T. gondii in cat fecal samples in our study in Mazandaran Province is very high. It seem that T. gondii infections in cats could be caused by the complicated environment of farms, accidental ingestion of undercooked food, and the presence of other intermediate hosts such as sheep and goats [3].
- It seems that the performance of high sensitivity molecular tests and the use of the RE gene marker, which is repeated about 200 times to 300 times throughout the T. gondii genome [34], was very effective for identifying the high prevalence of T. gondii in this study. Furthermore, other studies of this important zoonotic parasite in different human and animal groups in Mazandaran Province have confirmed its high prevalence. For example, the prevalence of T. gondii was 75.6% in women referred to a prenatal laboratory in Mazandaran [35], 77.4% in AIDS patients [36], and 58.8% in pregnant women [37]. According to a study conducted by Sharif et al. [38] in Mazandaran Province, the toxoplasmosis prevalence in sheep and goats was 35% and 30%, respectively, which is consistent with the high prevalence of T. gondii in cats. Geographical location and climatic conditions (including temperature and humidity) are important factors for the survival of oocysts; specifically, this parasite tends to be prevalent in areas with high humidity. The high prevalence of Toxoplasma infections in northern Iran may be due to the high humidity of 90% and moderate temperatures of 18-20°C [17].
- In conclusion, this is the first study in Iran to provide data related to presence of the Toxoplasmatinae subfamily in dog and cat fecal samples using PCR. The presence of T. gondii and N. caninum in cat and dog feces in northern Iran indicates a risk of transmission of toxoplasmosis and neosporosis infections. Additionally, the findings of this study suggest that public-health monitoring for the effective control of feces from cats and dogs and improved pet hygiene habits may be needed.
DISCUSSION
-
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare for this study.
-
FUNDING
None.
-
AUTHOR CONTRIBUTIONS
Conceptualization: AD, MS, SS. Data curation: AD, SS. Formal analysis: SAH. Funding acquisition: None. Methodology: LI, SAH, AA, MTR, TN. Project administration: AD, SS. Writing –original draft: LI, SAH, TN. Writing – review & editing: LI, SS, SAH, AA, MS, MTR, TN, AD.
NOTES
ACKNOWLEDGEMENTS
| Parasite | Primer | Size, bp | Annealing temperature, °C | Time, sec | Reference |
|---|---|---|---|---|---|
| N. caninum | NP21+: 5’-CCCAGTGCGTCCAATCCTGTAAC-3’ | 337 | 63 | 60 | [16] |
| NP6+: 5’-CTCGCCAGTCAACCTACGTCTTCT-3’ | |||||
| H. heydorni | JS4: 5’-CGAAATGGGAAGTTTTGTGAAC-3’ | 267 | 65 | 60 | [13] |
| JS5: 5’-CAGCAGCTAGATACGTAGA-3’ | |||||
| T. gondii | TOX4: 5’-CGCTGCAGGGAGGAAGACGAAAGTTG-3’ | 529 | 55 | 30 | [17] |
| TOX5: 5’-CGCTGCAGACACAGTGCATCTGGATT-3’ | |||||
| H. hammondi | Hham34F: 5’- ATCCCATTCCGGCTTCAGTCTTTC-3’ | 282 | 60 | 60 | [18] |
| Hham3R: 5’-ACAGCGGAGCCGAAGTTGGTTTR-3’ | |||||
| B. besnoiti | ITS-1F: 5’-TGACATTTAATAACAATCAACCCTT-3’ | 230 | 58 | 90 | [19] |
| ITS-1R: 5’-GGTTTGTATTAACCAATCCGTGA-3’ |
| Animals | T. gondii | N. caninum | Hammondia spp. | B. besnoiti |
|---|---|---|---|---|
| Dog | 1/120 (0.8) | 2/120 (1.7) | 0/120 (0.0) | 0/120 (0.0) |
| Cat | 18/80 (22.5) | 0/80 (0.0) | 0/80 (0.0) | 0/80 (0.0) |
- 1. Mugridge NB, Morrison DA, Heckeroth AR, Johnson AM, Tenter AM. Phylogenetic analysis based on full-length large subunit ribosomal RNA gene sequence comparison reveals that Neospora caninum is more closely related to Hammondia heydorni than to Toxoplasma gondii. Int J Parasitol 1999;29:1545-1556.ArticlePubMed
- 2. Webster JP. Rats, cats, people and parasites: the impact of latent toxoplasmosis on behaviour. Microbes Infect 2001;3:1037-1045.ArticlePubMed
- 3. Dubey JP. Toxoplasmosis of animals and humans. Boca Raton: CRC Press; 2016. p 77-82.
- 4. Schares G, Pantchev N, Barutzki D, Heydorn AO, Bauer C, Conraths FJ. Oocysts of Neospora caninum, Hammondia heydorni, Toxoplasma gondii and Hammondia hammondi in faeces collected from dogs in Germany. Int J Parasitol 2005;35:1525-1537.PubMed
- 5. Dubey JP, Hill DE, Lindsay DS, Jenkins MC, Uggla A, Speer CA. Neospora caninum and Hammondia heydorni are separate species/organisms. Trends Parasitol 2002;18:66-69.ArticlePubMed
- 6. Cavalcante GT, Aguiar DM, Chiebao D, Dubey JP, Ruiz VL, Dias RA, et al. Seroprevalence of Toxoplasma gondii antibodies in cats and pigs from rural Western Amazon, Brazil. J Parasitol 2006;92:863-864.ArticlePubMed
- 7. Gondim LF. Neospora caninum in wildlife. Trends Parasitol 2006;22:247-252.ArticlePubMed
- 8. Mehlhorn H, Heydorn AO. Neospora caninum: is it really different from Hammondia heydorni or is it a strain of Toxoplasma gondii? An opinion. Parasitol Res 2000;86:169-178.ArticlePubMed
- 9. Cortes H, Leitão A, Gottstein B, Hemphill A. A review on bovine besnoitiosis: a disease with economic impact in herd health management, caused by Besnoitia besnoiti (Franco and Borges,). Parasitology 2014;141:1406-1417.ArticlePubMed
- 10. Dubey JP, Sreekumar C. Redescription of Hammondia hammondi and its differentiation from Toxoplasma gondii. Int J Parasitol 2003;33:1437-1453.ArticlePubMed
- 11. Dubey JP, Zarnke R, Thomas NJ, Wong SK, Van Bonn W, Briggs M, et al. Toxoplasma gondii, Neospora caninum, Sarcocystis neurona, and Sarcocystis canis-like infections in marine mammals. Vet Parasitol 2003;116:275-296.ArticlePubMed
- 12. Walsh CP, Duncan RB, Zajac AM, Blagburn BL, Lindsay DS. Neospora hughesi: experimental infections in mice, gerbils, and dogs. Vet Parasitol 2000;92:119-128.ArticlePubMed
- 13. Slapeta JR, Koudela B, Votýpka J, Modrý D, Horejs R, Lukes J. Coprodiagnosis of Hammondia heydorni in dogs by PCR based amplification of ITS 1 rRNA: differentiation from morphologically indistinguishable oocysts of Neospora caninum. Vet J 2002;163:147-154.ArticlePubMed
- 14. Slapeta JR, Modrý D, Kyselová I, Horejs R, Lukes J, Koudela B. Dog shedding oocysts of Neospora caninum: PCR diagnosis and molecular phylogenetic approach. Vet Parasitol 2002;109:157-167.ArticlePubMed
- 15. Silva RC, Caffaro K, Paula CL, Risseti RM, Langoni H, Megid J, et al. An atypical Toxoplasma gondii genotype in a rural Brazilian dog co-infected with Leishmania (Viannia) braziliensis. Rev Soc Bras Med Trop 2015;48:224-227.ArticlePubMed
- 16. Müller N, Zimmermann V, Hentrich B, Gottstein B. Diagnosis of Neospora caninum and Toxoplasma gondii infection by PCR and DNA hybridization immunoassay. J Clin Microbiol 1996;34:2850-2852.ArticlePubMedPMC
- 17. Hosseini SA, Golchin E, Sharif M, Sarvi S, Ahmadpour E, Rostamian A, et al. A serological investigation and genotyping of Toxoplasma gondii among Iranian blood donors indicates threat to health of blood recipients. Transfus Apher Sci 2020;59:102723.ArticlePubMed
- 18. King JS, Brown GK, Jenkins DJ, Ellis JT, Fleming PJ, Windsor PA, et al. Oocysts and high seroprevalence of Neospora caninum in dogs living in remote Aboriginal communities and wild dogs in Australia. Vet Parasitol 2012;187:85-92.ArticlePubMed
- 19. Cortes HC, Reis Y, Gottstein B, Hemphill A, Leitão A, Müller N. Application of conventional and real-time fluorescent ITS1 rDNA PCR for detection of Besnoitia besnoiti infections in bovine skin biopsies. Vet Parasitol 2007;146:352-356.ArticlePubMed
- 20. Lindsay DS, Dubey JP. In vitro development of Neospora caninum (Protozoa: Apicomplexa) from dogs. J Parasitol 1989;75:163-165.ArticlePubMed
- 21. McAllister M, Wills RA, McGuire AM, Jolley WR, Tranas JD, Williams ES, et al. Ingestion of Neospora caninum tissue cysts by Mustela species. Int J Parasitol 1999;29:1531-1536.ArticlePubMed
- 22. Reid AJ, Vermont SJ, Cotton JA, Harris D, Hill-Cawthorne GA, Könen-Waisman S, et al. Comparative genomics of the apicomplexan parasites Toxoplasma gondii and Neospora caninum: Coccidia differing in host range and transmission strategy. PLoS Pathog 2012;8:e1002567.ArticlePubMedPMC
- 23. Barratt J, Al Qassab S, Reichel MP, Ellis JT. The development and evaluation of a nested PCR assay for detection of Neospora caninum and Hammondia heydorni in feral mouse tissues. Mol Cell Probes 2008;22:228-233.ArticlePubMed
- 24. Hill DE, Liddell S, Jenkins MC, Dubey JP. Specific detection of Neospora caninum oocysts in fecal samples from experimentally-infected dogs using the polymerase chain reaction. J Parasitol 2001;87:395-398.ArticlePubMed
- 25. Razmi GR. Survey of dogs’parasites in Khorasan Razavi Province, Iran. Iranian J Parasitol 2009;4:48-54.
- 26. Ghafarifar F, Sabevarinejad G, Dalimi A, Forouzandeh-Moghadam M. Molecular detection of Neospora caninum from naturally infected dogs in Lorestan province, West of Iran. Arch Razi Inst 2014;69:185-190.
- 27. Jie HJ, Yu M, Fen YY, Mei GY, Yan Y, Esch GW, et al. First isolation of Hammondia heydorni from dogs in China. Vet Parasitol 2013;197:43-49.ArticlePubMed
- 28. Li J, He P, Yu Y, Du L, Gong P, Zhang G, et al. Detection of Neospora caninum-DNA in feces collected from dogs in Shenyang (China) and ITS1 phylogenetic analysis. Vet Parasitol 2014;205:361-364.ArticlePubMed
- 29. Etheredge GD, Michael G, Muehlenbein MP, Frenkel JK. The roles of cats and dogs in the transmission of Toxoplasma infection in Kuna and Embera children in eastern Panama. Rev Panam Salud Publica 2004;16:176-186.ArticlePubMed
- 30. Mancianti F, Nardoni S, Ariti G, Parlanti D, Giuliani G, Papini RA. Cross-sectional survey of Toxoplasma gondii infection in colony cats from urban Florence (Italy). J Feline Med Surg 2010;12:351-354.ArticlePubMed
- 31. Berger-Schoch AE, Herrmann DC, Schares G, Müller N, Bernet D, Gottstein B, et al. Prevalence and genotypes of Toxoplasma gondii in feline faeces (oocysts) and meat from sheep, cattle and pigs in Switzerland. Vet Parasitol 2011;177:290-297.ArticlePubMed
- 32. Jokelainen P, Simola O, Rantanen E, Näreaho A, Lohi H, Sukura A. Feline toxoplasmosis in Finland: cross-sectional epidemiological study and case series study. J Vet Diagn Invest 2012;24:1115-1124.ArticlePubMedPDF
- 33. Dubey JP, Darrington C, Tiao N, Ferreira LR, Choudhary S, Molla B, et al. Isolation of viable Toxoplasma gondii from tissues and feces of cats from Addis Ababa, Ethiopia. J Parasitol 2013;99:56-58.ArticlePubMed
- 34. Edvinsson B, Lappalainen M, Evengård B; ESCMID Study Group for Toxoplasmosis. Real-time PCR targeting a 529-bp repeat element for diagnosis of toxoplasmosis. Clin Microbiol Infect 2006;12:131-136.ArticlePubMed
- 35. Âjami A, Sharif M, Saffar M, Zyaee H. Serological study of toxoplasmosis in women referred to medical health laboratory before marriage, Mazandaran, 2000. J Mazandaran Univ Med Sci 2001;11:51-56 (Persian).
- 36. Daryani A, Sharif M, Meigouni M. Seroprevalence of IgG and IgM anti-Toxoplasma antibodies in HIV/AIDS patients, northern Iran. Asian Pac J Trop Med 2011;4:271-274.ArticlePubMed
- 37. Hoseini SA, Dehgani N, Sharif M, Daryani A, Gholami S, Ebrahimi F, et al. Serological survey of toxoplasmosis in pregnant women. J Mazandaran Univ Med Sci 2014;24:146-150 (Persian).
- 38. Sharif M, Gholami Sh, Ziaei H, Daryani A, Laktarashi B, Ziapour SP, et al. Seroprevalence of Toxoplasma gondii in cattle, sheep and goats slaughtered for food in Mazandaran province, Iran, during 2005. Vet J 2007;174:422-424.ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- Prevalence of Toxoplasma gondii in Endangered Wild Felines (Felis silvestris and Lynx pardinus) in Spain
Pablo Matas Méndez, Isabel Fuentes Corripio, Ana Montoya Matute, Begoña Bailo Barroso, Rebeca Grande Gómez, Ariadna Apruzzese Rubio, Francisco Ponce Gordo, Marta Mateo Barrientos
Animals.2023; 13(15): 2488. CrossRef - Endoparasites of European Wildcats (Felis silvestris) in Greece
Anastasia Diakou, Despina Migli, Dimitris Dimzas, Simone Morelli, Angela Di Cesare, Dionisios Youlatos, Petros Lymberakis, Donato Traversa
Pathogens.2021; 10(5): 594. CrossRef

 KSE
KSE
 PubReader
PubReader ePub Link
ePub Link Cite
Cite