Articles
- Page Path
- HOME > Epidemiol Health > Volume 44; 2022 > Article
-
Original Article
High frequency of colonization by extended-spectrum beta-lactamase-producing Gram-negative bacilli in hemodialysis patients and their household contacts in Colombia: dissemination between the community and the hospital -
Daniela Montoya-Urrego1
 , Sara Tellez-Carrasquilla1
, Sara Tellez-Carrasquilla1 , Johanna M. Vanegas1,2
, Johanna M. Vanegas1,2 , Judy Natalia Jiménez Quiceno1
, Judy Natalia Jiménez Quiceno1
-
Epidemiol Health 2022;44:e2022069.
DOI: https://doi.org/10.4178/epih.e2022069
Published online: August 27, 2022
1Grupo de Investigación en Microbiología Básica y Aplicada (MICROBA), Escuela de Microbiología, Universidad de Antioquia, Medellín, Colombia
2Grupo de Investigación en Salud Pública, Escuela de Ciencias de la Salud, Universidad Pontificia Bolivariana, Medellín, Colombia
- Correspondence: Judy Natalia Jiménez Quiceno Grupo de Investigación en Microbiología Básica y Aplicada (MICROBA), Escuela de Microbiología, Universidad de Antioquia, Calle 67 No. 53-108, Medellín 050010, Colombia E-mail: jnatalia.jimenez@udea.edu.co
© 2022 Korea Disease Control and Prevention Agency.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Abstract
-
OBJECTIVES
- Increasing colonization by beta-lactam-resistant Gram-negative bacilli (BR-GNB) represents a risk for infections and bacterial resistance spread, both in hospitals and the community. Hemodialysis patients and their household contacts regularly transit between these environments. This study investigated the clinical and epidemiological characteristics of BR-GNB colonization in hemodialysis patients and their household contacts, as well as the genetic relationship between their isolates.
-
METHODS
- A cross-sectional study was conducted on hemodialysis patients at a hospital-associated dialysis center in Medellín, Colombia and their household contacts. Clinical and epidemiological information was collected. Colonization was assessed from stool or rectal swab samples. Bacterial identification and susceptibility were determined using chromogenic media and Vitek-2. Molecular characterization included beta-lactamase detection by polymerase chain reaction, multiple-locus sequence typing (MLST), pulsed-field gel electrophoresis, and identification of Escherichia coli phylogroups by the Clermont protocol.
-
RESULTS
- This study included 36 hemodialysis patients and 90 household contacts. Colonization by BR-GNB occurred in 58.3% of patients and 22.2% of household contacts. The main beta-lactamase detected was CTX-M group-1 (40.5%). In 3 of the 9 homes that had more than 1 colonized individual, a genetic relationship was found. MLST showed a high diversity in E. coli isolates, and the most frequent phylogroups were B1 and B2.
-
CONCLUSIONS
- These results show a high frequency of colonization and the presence of potentially pathogenic BR-GBN both in hospitals and the community. This highlights the importance of populations who move between those 2 environments, and the need to prevent the spread of bacterial resistance outside hospitals.
- Beta-lactam-resistant Gram-negative bacilli (BR-GNB) have emerged as important pathogens that can cause a variety of healthcare-associated or community-acquired infections, and have been on the rise, according to the latest Centers for Disease Control and Prevention (CDC) reports [1]. Infections by this type of bacteria have serious implications in terms of increased mortality, longer hospital stays due to a delay in adequate treatment, and extra costs for health systems [1-3].
- Hemodialysis patients present specific clinical characteristics that favor high frequencies of colonization by BR-GNB, even higher than those reported for other bacteria of clinical importance such as Staphylococcus aureus, therefore increasing the risk of infection [4]. In addition, these patients constantly circulate between hospital and community environments, so they act as carriers of these bacteria for long periods, facilitating their transmission to people with whom they interact closely, such as members of their family and their contacts within the community [5,6]. Likewise, hemodialysis patients share a link of care with their household contacts since they need constant accompaniment and assistance. Therefore, their household contacts also interact with the hospital environment and share spaces, objects, and habits with patients at home, which can facilitate the transmission of bacteria [7]. At the same time, different social, economic, epidemiological, cultural, and environmental factors converge in this population, which can favor colonization by resistant bacteria from community sources.
- Accordingly, hemodialysis patients and their household contacts are a model population to understand the behavior of the colonization of BR-GNB between the hospital environment and the community. Taking into account that Colombia is an endemic country for BR-GNB, and that, particularly in Medellín, colonization by these microorganisms in hemodialysis patients has been reported in up to 52.7% [4], this study aimed to determine the clinical and epidemiological characteristics of colonization by BR-GNB in hemodialysis patients and their household contacts, in order to reach an understanding of the behavior of colonization between the hospital and the community, and to identify prevention and control strategies to reduce the spread of resistant bacteria.
INTRODUCTION
- Study population
- A cross-sectional study was conducted among hemodialysis patients and their household contacts between June 2019 and May 2020. The patients were treated at a dialysis center associated with a tertiary hospital in Medellín, which cares for approximately 350 patients. As selection criteria, hemodialysis patients had to share a home with at least 1 person; household contacts were defined as individuals who lived in the same house as the patient for a period equal to or greater than 6 months. For minors, consent was provided by their parents or legal guardians. Finally, if pets, such as cats and dogs, lived in the homes, they were screened.
- Based on a total of 106 eligible hemodialysis patients, 70 were excluded for the following reasons: (1) 3 lived in temporary shelters or convents, (2) 6 lived alone, (3) 23 could not be contacted, (4) 29 refused to participate, and (5) 9 felt uncomfortable with sample collection. Meanwhile, of 146 eligible household contacts, 56 were excluded, since (1) 23 refused to participate and (2) 33 felt uncomfortable with the sample collection. A total of 126 participants distributed among 38 homes were included: 36 hemodialysis patients and 90 of their household contacts (Supplementary Material 1).
- Data collection
- Hemodialysis patients were contacted by telephone from a cohort of a previous study in our research group, who presented previous colonization [4]. A visit to each home was arranged and, once informed consent had been provided, the clinical and epidemiological information was obtained using a form designed for this purpose. The information included general characteristics, clinical information, self-reports of hand-washing and eating habits (times per week). If there were pets in the home, the pets’ information and clinical history were included.
- Screening for beta-lactam-resistant Gram-negative bacilli colonization
- Colonization by BR-GNB was evaluated based on stool samples of the patients and their household contacts, and rectal swabs from pets. Each sample was enriched in trypticase soy broth and incubated for 12 hours to 18 hours at 37°C. Subsequently, 100 µL of broth was seeded in the chromogenic media ChromID-ESBL and ChromID-CARBA (bioMérieux, Marcy-l’Étoile, France) for the selection of extended-spectrum beta-lactamase (ESBL)-producing and carbapenem-resistant Gram-negative bacilli [8,9]. To increase the sensitivity in the detection of carbapenem-resistant Gram-negative bacilli, the protocol validated by the CDC was also used, where the sample was enriched by adding an ertapenem disk (10 µg) and seeded on McConkey agar [10]. Bacterial identification and susceptibility testing were performed using the automated Vitek-2 system (bioMérieux). The antibiotics evaluated were amikacin, ampicillin/sulbactam, cefepime, cefoxitin, ceftazidime, ceftriaxone, ciprofloxacin, doripenem, ertapenem, gentamicin, imipenem, meropenem, piperacillin/tazobactam, and tigecycline, according to the Clinical and Laboratory Standards Institute cut-off points [11].
- Molecular detection of resistance mechanisms to beta-lactam antibiotics
- DNA of the isolates was extracted using the Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Detection of beta-lactamases was carried out using polymerase chain reaction (PCR) amplification of genes that encode for CTX-M-G1, CTX-M-G2, CTX-M-G9, CTX-M-G8/25, TEM, and SHV [12]. Additionally, in carbapenem-resistant Gram-negative bacilli, the presence of genes encoding carbapenemase types KPC, IMP, VIM, NDM, and OXA-48 was evaluated by PCR, according to the protocols previously described and standardized in the laboratory [13,14].
- Molecular typing of isolates
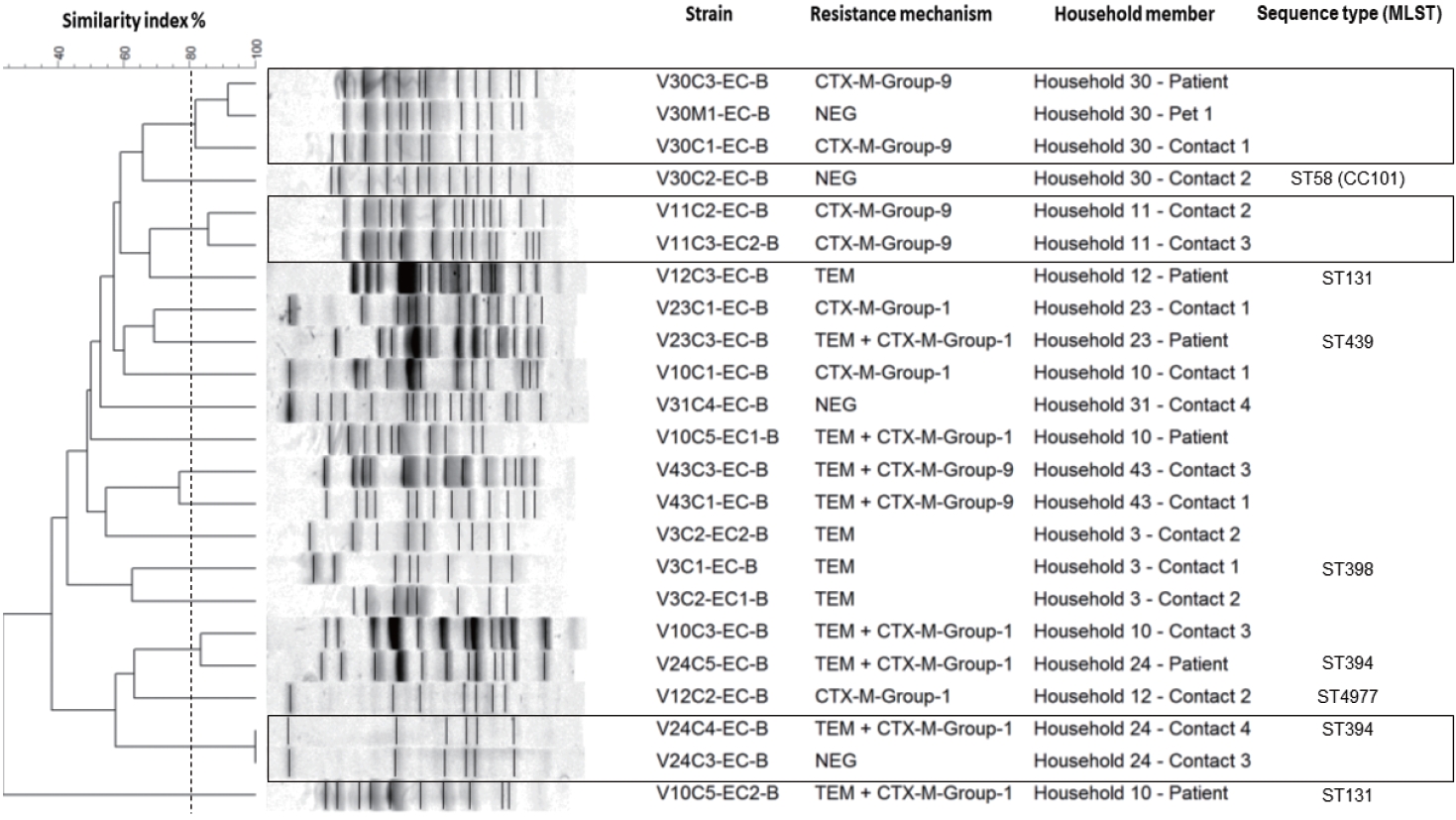
- In the event that at least 2 household members were colonized in the same home, the genetic relatedness of isolates from hemodialysis patients, household contacts, and pets was determined using pulsed-field gel electrophoresis (PFGE), which was performed using the XbaI restriction enzyme (Thermo Scientific, Waltham, MA, USA) [15]. The cluster analysis was performed using BioNumerics software version 6.0, and dendrograms were generated by the unweighted pair group method using average linkages. A similarity cutoff of 80% was used to define genetically related strains.
- Additionally, multiple-locus sequence typing (MLST) was performed on a representative sample of the isolates, according to the group generated by PFGE [16]. The sequence type and the clonal complex were assigned using the web database (https://pubmlst.org/). Finally, all Escherichia coli isolates were typified using the Clermont method to establish the phylogroups that represented the highest clinical risk in colonized people. For this, quadruplex PCR and 2 simple PCRs were performed to recognize the 7 defined phylogroups (A, B1, B2, C, D, E, and F) according to the previously described protocol [17].
- Statistical analysis
- Categorical variables were described with absolute and relative frequencies. Continuous variables were expressed with the mean and standard deviation or median and interquartile range (IQR), according to the assumption of normality. To determine the factors potentially associated with colonization in hemodialysis patients, a multivariate analysis using a generalized linear model for a Poisson distribution with a log-link function and a robust estimator of variance was performed. For household contacts, as there may be several per home, it was necessary to adjust for the cluster effect, so a generalized estimating equation model was performed for a Poisson distribution with a log-link function, assuming an exchangeable correlation and a robust estimator of variance. Each exposure was analyzed in a different model, adjusted for age; the association measures were prevalence ratios (PRs), expressed with their corresponding confidence interval (95% CI) and p-value. The statistical analysis was performed using Stata version 14.0 (StataCorp., College Station, TX, USA).
- Ethics statement
- This study was approved by the Committee for Bioethics in Humans of the University of Antioquia (CBEIH-SIU), approval act 18-35-820, and by the Ethics Committee for Experimentation with Animals of the University of Antioquia (CEEA) with the minutes of session No. 136. All participants provided in formed consent.
MATERIALS AND METHODS
- Most of the patients were men (55.6%, n= 20), the median age was 61 years old (IQR, 47-71), and the median number of household contacts per patient was 3 (IQR, 2-4). The main comorbidities were arterial hypertension (80.6%, n= 29) and diabetes mellitus (33.3%, n= 12). The clinical history revealed that 50% of patients (n= 18) had been hospitalized or undergone surgery in the last year, and 50% (n= 18) consumed antibiotics in the same period. Furthermore, 16.7% (n= 6) of patients had traveled in the last year, mainly locally.
- Regarding the household contacts, the majority were women (71.1%, n= 64), and their median age was 39 years (IQR, 17-59). Their clinical histories revealed that 13.3% (n= 12) of the household contacts were hospitalized in the last year, and 30% (n= 27) consumed antibiotics in the same period. The main comorbidities reported were arterial hypertension (16.7%, n= 15) and diabetes mellitus (12.2%, n= 11). In addition, 31.1% of household contacts (n= 28) traveled in the last year. Other clinical and epidemiological data are summarized in Table 1.
- Colonization by beta-lactam-resistant Gram-negative bacilli
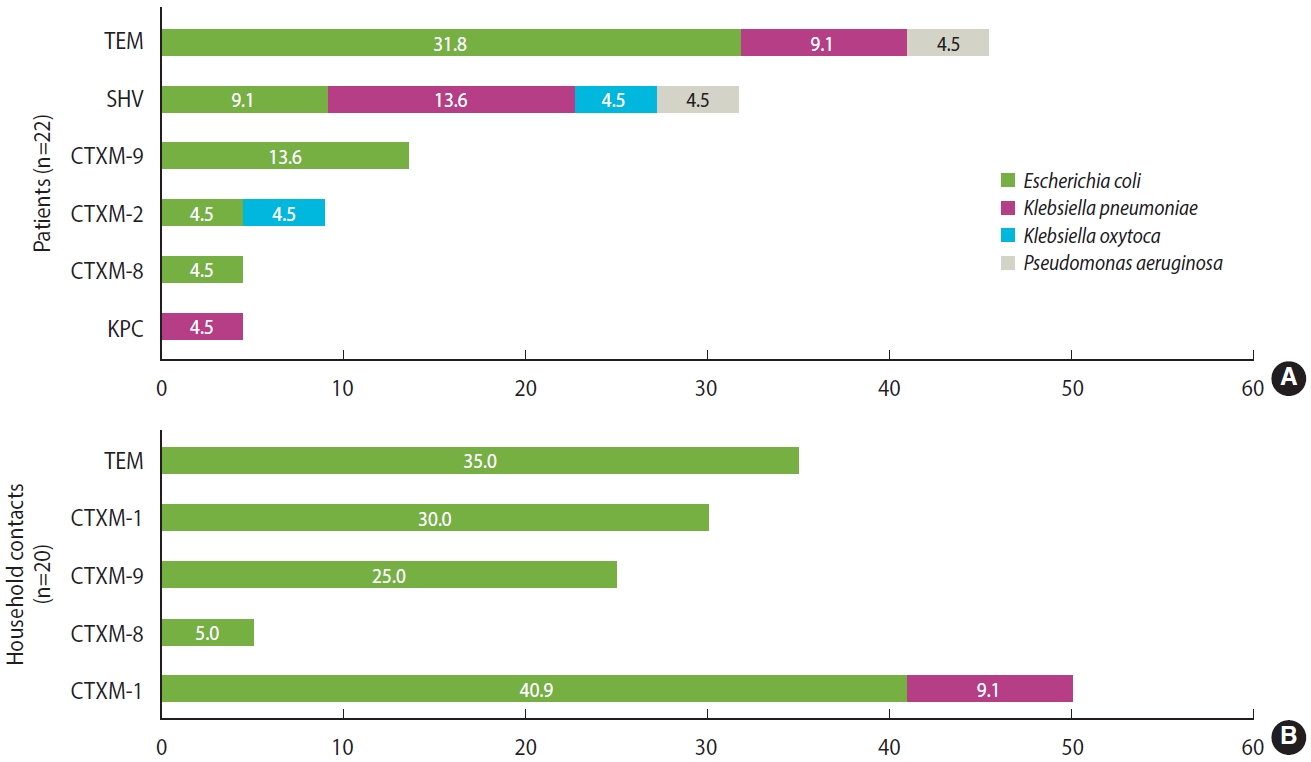
- In general, colonization by BR-GNB was found in 32.5% (n= 41) of the participants. In hemodialysis patients, it was present in 58.3% of the patients (n= 21/36); 22 isolates were obtained, most of which were E. coli (77.3%, n= 17), followed by Klebsiella pneumoniae (13.6%, n= 3), Klebsiella oxytoca and Pseudomonas aeruginosa (4.5%, n= 1). Of the 22 isolates, 19 ESBL carriers were detected (86.4%), 2 were resistant to carbapenems, and 1 isolate was resistant to third-generation cephalosporins without the presence of ESBL but with a TEM-type beta-lactamase.
- Among the household contacts, colonization was found in 22.2% (n= 20/90); 20 isolates were obtained, most of them E. coli (95%, n= 19) and 1 isolate of P. aeruginosa. Of the 20 isolates, 16 ESBL carriers were detected (80%), 1 was resistant to carbapenems, and 3 to third-generation cephalosporins without the presence of ESBL, but with a TEM-type beta-lactamase.
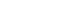
- Of 38 homes evaluated, colonized people were found in 26 (68.4%), of which 13 homes had only colonized patients (50.0%), in 19.2% (n= 5) only colonized household contacts were found, and in 30.7% of homes (n= 8), colonized patients and household contacts were found (Figure 1). Colonization by E. coli of 2 or more members of the same home was detected in 23.6% (n= 9) of homes. Additionally, of the 25 pets evaluated (10 canines and 15 felines), colonization by BR-GNB was found in 2 dogs; both isolates were E. coli.
- Mechanisms of resistance to beta-lactams
- Regarding resistance mechanisms, 83.3% (n= 35/42) of all isolates carried some type of ESBL. In hemodialysis patients, 50% of isolates carried ESBL CTX-M group 1, which includes CTX-M-1, CTX-M-3, and CTX-M-15 variants (40.9% E. coli and 9.1% K. pneumoniae), followed by CTX-M-9 with 13.6% (n= 3, E. coli); TEM and SHV beta-lactamases were also identified in a significant proportion of isolates (45.4% and 31.7%, respectively). Furthermore, 50% of the isolates carried at least 2 beta-lactamases simultaneously. In household contacts, the most common beta-lactamase was TEM (35%, n= 7), followed by ESBL CTX-M-group 1 (30%, n= 6), both detected in isolates of E. coli. Similarly, 20% of the isolates carried 2 beta-lactamases simultaneously. Regarding carbapenemases, KPC could be identified from a K. pneumoniae isolate. Importantly, NDM, OXA-48, VIM, or IMP type carbapenemases were not detected. All detected beta-lactamases are shown in Figure 2.
- Molecular typing of isolates
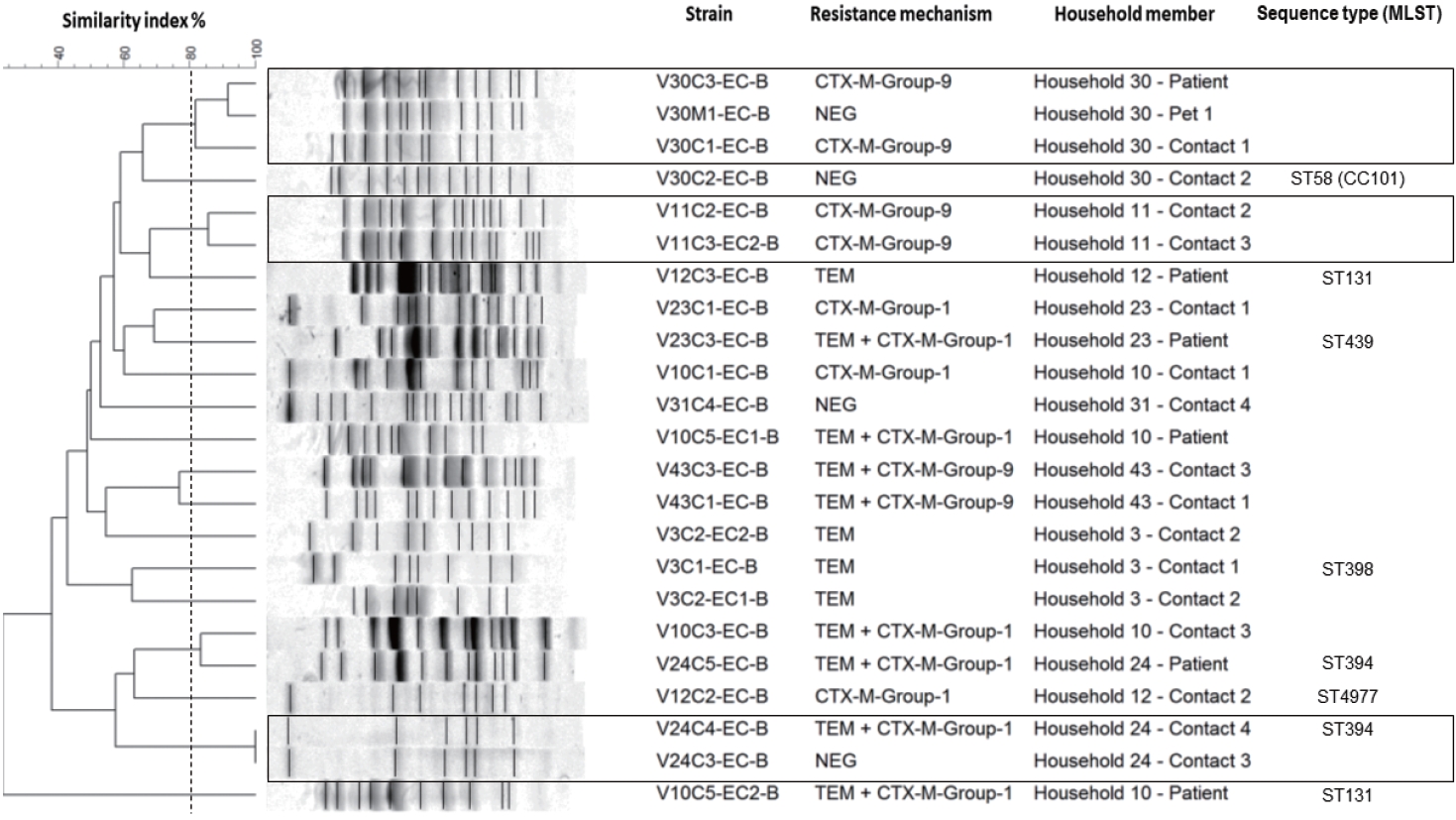
- The genetic relatedness was evaluated in 9 homes, in which 2 or more inhabitants were colonized by E. coli (Figure 1). In total, 23 isolates were evaluated by PFGE, resulting in a genetic relationship between isolates from different members in 3 homes (33.3%); however, extensive diversity was identified in the E. coli isolates (Figure 3). It should be noted that in 1 family, there was a genetic relationship between the patient, 1 of their household contacts, and their pet (canine). Additionally, MLST was performed on 12 E. coli isolates, and the results showed the presence of ST131 and ST394 (2 isolates each), and ST398, ST4977, ST8420, ST349, ST101, ST58, ST457, and ST6011 (1 isolate each).
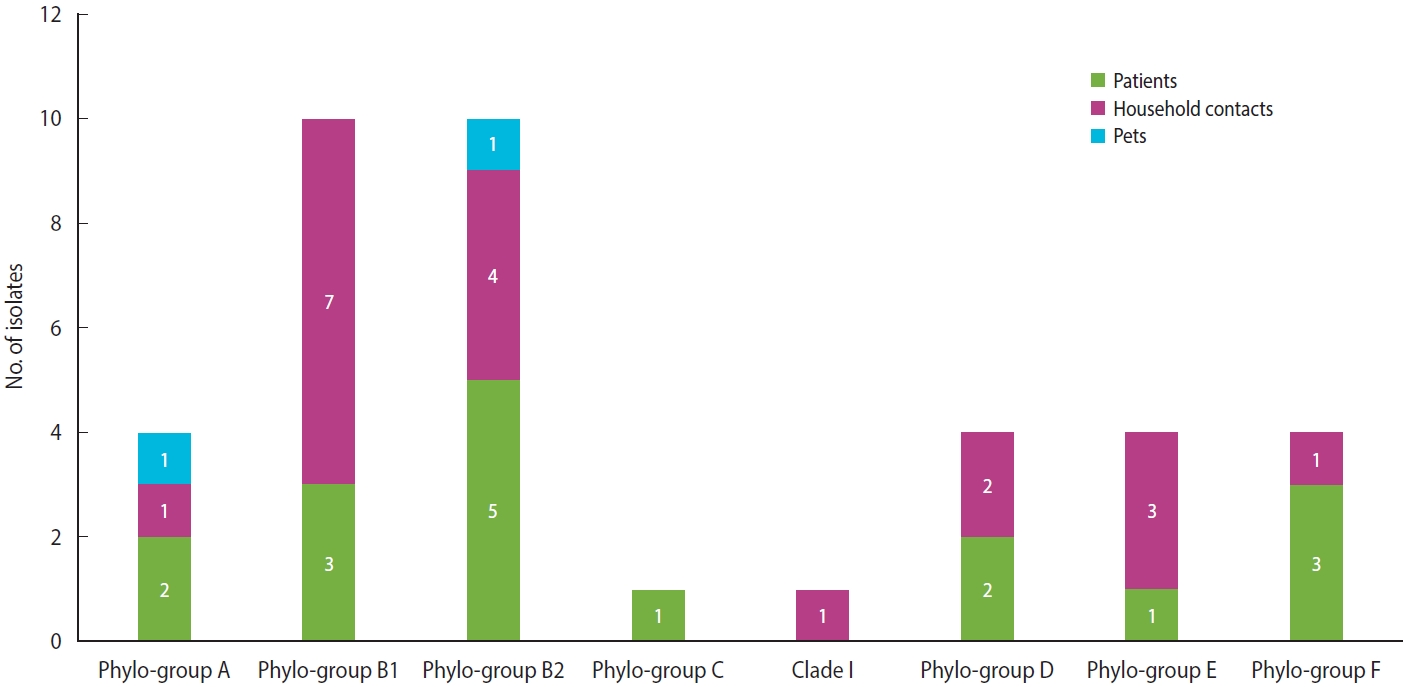
- Regarding the classification by the Clermont scheme, 38 isolates of E. coli were analyzed, where the main phylogroups identified were B1 and B2, each with 26.3% (n= 10), followed by phylogroups A, D, E, and F, each with 10.5% (n= 4). Finally, phylogroup C and clade 1 were identified, each with 2.6% (n= 1) (Figure 4).
- Factors associated with colonization
- The potential factors associated with colonization were analyzed for ESBL-producing Gram-negative bacilli, and the results are presented in Tables 2 and 3, for hemodialysis patients and household contacts, respectively. Colonization was positively associated with not washing hands (PR, 1.89; 95% CI, 1.38 to 2.58) and the presence of comorbidities, such as cancer (PR, 2.57; 95% CI, 1.42 to 4.63). However, washing hands after using the toilet (PR, 0.46; 95% CI, 0.30 to 0.71) acted as a protective factor. Other factors such as previous antibiotic consumption did not show associations with colonization by ESBL-producing Gram-negative bacilli.
RESULTS
- The results of this study demonstrated a high frequency of colonization not only in hemodialysis patients, but also in their household contacts. Although several studies have evaluated colonization by BR-GNB, this work started from a high-risk group and took into account a population that constantly circulates between hospital and the community. This allowed us to refine our understanding of the behavior of colonization between community and hospital environments and to observe the role of homes as a possible reservoir and disseminator of this type of bacteria.
- The frequency of BR-GNB colonization in hemodialysis patients was similar to that reported in a previous study carried out in hemodialysis patients from the same renal unit, where 41.2% and 11.5% of patients were colonized by ESBL-producing and carbapenem-resistant Gram-negative, respectively [4]. Our results exceed the prevalence of BR-GNB reported in other studies, such as Wendt et al. [18], of a kidney unit in Germany, where colonization by multi-resistant Gram-negative bacteria was 10.4%. This difference could be explained by the high hygiene standards and the strict antibiotic use policies in that institution.
- Regarding the household contacts of hemodialysis patients, there are few studies evaluating colonization by BR-GNB in this population. However, the frequency reported in this study exceeds the values reported for the general community, which are between 9.8% and 14% according to some studies [19-21]. In studies carried out in relatives of other types of patients, such as patients with hemolytic uremic syndrome, a frequency of colonization by E. coli of 12% was reported [22], while Adler et al. [23] found ESBL-producing enterobacteria in 9.1% of relatives of patients hospitalized in rehabilitation centers. One of the reasons for these differences may be that the household contacts of hemodialysis patients are in constant transit between the hospital and the community due to their bond of care with the patients, which favors contact with the hospital environment. In addition, the frequencies of colonization are also affected by geographic location, and Colombia is considered an endemic country for ESBL-producing Gram-negative bacilli [24]. Nonetheless, the consumption of antibiotics in the household contacts was very high, which may favor the selection pressure for resistant bacteria in this population.
- The importance of colonization by BR-GNB in the general community lies in the fact that these people can be the main source of transport and dissemination of these microorganisms, so characterizing this population men it possible to know the behavior of resistance at the local level [25]. Accordingly, the relatives of hemodialysis patients can transport these resistant bacteria in the community and in the hospital; this contact with health institutions by accompanying patients has been described as a possible cause that strengthens the community transmission of ESBL-producing Enterobacteriaceae [20]. However, the percentage of colonization by BR-GNB in animals is similar to the data reported in a study in New Zealand, where the 6.4% of dogs and cats were colonized by ESBL-producing E. coli, which indicates that companion animals can also be reservoirs and acquire resistant bacteria in homes or veterinary clinics [26].
- The presence of resistant bacteria in the community makes this environment an important source of acquisition of these microorganisms, not only in homes but also in places where there is frequent contact with other people or with potentially contaminated surfaces, such as supermarkets, public transport, and schools. In this sense, it is worth highlighting the importance of designing and implementing educational strategies that improve knowledge about bacterial resistance and the proper use of antibiotics in the general community, based on the particular needs and epidemiological behavior of resistance in different populations.
- The PFGE and MLST results show that the high diversity of isolates in our population and confirm E. coli as a microorganism with high genetic plasticity. Likewise, it suggests a strong selection pressure in different places that behave as sources of colonization, which can occur at the hospital level or from community spaces, since our population is in constant transit between the 2 environments. Once colonized, patients and their household contacts become carriers of this type of bacteria and can help it to spread.
- However, the genetic relationship found in isolates from different members in 3 families confirms that bacteria can be shared in homes between inhabitants, including animals, and shows homes as potential reservoirs for bacterial resistance. This has been described in other populations, which place the relationship between colonizing strains of ESBL-producing E. coli between members of the same household between 8% and 27%. An exchange of bacteria has also been seen between domestic animals and their caregivers, with a relationship reported in between 25% and 27% of cases [27,28]. This highlights the importance of taking into account homes, household contacts, and companion animals in strategies to control the spread of resistant bacteria.
- Concerning E. coli, classification by the Clermont system made it possible to differentiate commensal strains from intestinal and extraintestinal pathogenic variants; this is important to stablish a relationship between colonizing strains and disease, which is information of vital importance in this type of patient [29,30]. In this study, the most frequent E. coli phylogroups were B1, which has been identified as a cause of infections in dialysis-dependent patients [31], and B2, which has been considered the main phylogroup worldwide in causing extraintestinal disease [32] and, in turn, has been documented as one of the main colonizers of the gastrointestinal tract of patients with other chronic diseases [33,34], being related to ST131, which is characterized by presenting a resistance profile to beta-lactam antibiotics [35]. Likewise, the isolates from pets in our study were classified into phylogroups A and B2, coinciding with previous reports, where dogs were considered potential transmitters of pathogenic E. coli to their caregivers [36].
- Regarding the factors associated with colonization, good hygiene practices have been documented as protective factors for colonization by resistant bacteria, which is consistent with the data obtained in this study, where the absence of handwashing was related to colonization [37,38]. Similarly, having chronic diseases such as cancer and using antibiotics are among the most frequently described risk factors for colonization by ESBL-GNB [39].
- Finally, regarding the resistance mechanisms detected, most of the isolates were E. coli carriers of ESBL, especially CTX-M-G1. This agrees with what has been reported in previous studies of hemodialysis patients in Colombia, where CTX-M-G1 was present in 47.9% of the E. coli isolates evaluated [40]. In general, beta-lactamases of the CTX-M type are widely distributed worldwide and are endemic in Latin America, North America, and Asia [41], so it is common to find microorganisms with these enzymes colonizing and infecting different population groups throughout the world [42-45].
- Regarding to carbapenem-resistant Gram-negative bacilli, other studies have reported from 2.4% to 4.2% of isolates carrying KPC in patients with chronic kidney disease. The difference from our study may be because this work is cross-sectional, but colonization can act intermittently, which could have influenced the results [45,46]. Nonetheless, carbapenemases were not detected in P. aeruginosa isolates; however, their resistance may be due to a combination of intrinsic resistance with modifications in the expression of porins or expulsion pumps. It should be noted that various reports point to an increase in BR-GNB, especially carriers of ESBL in the community, which shows the importance of continuing with studies aimed at knowing the real magnitude of colonization in this environment and the factors associated with it to establish more effective intervention strategies.
- In conclusion, the results of this study showed that there was a high frequency of colonization by BR-GNB both in hemodialysis patients and in their household contacts, confirming the presence of this type of bacteria both in people with contact with the hospital environment and in the community. Likewise, the most frequently isolated microorganism was ESBL-producing E. coli, and it was confirmed that the strains circulating in our population have pathogenic potential since most belonged to the B1 and B2 phylogroups. The diversity observed also suggests that, within the community, there are different sources from which these bacteria can be acquired, which implies that surveillance of bacterial resistance should be strengthened at the community level, and that preventative measures should be implemented. As for limitations and perspectives, this population could not be followed, which could improve the understanding of the behavior and duration of colonization by BR-GNB. Likewise, studies involving whole-genome sequencing could clarify the directionality of transmission between patients and household contacts, which would help direct more assertive control strategies.
DISCUSSION
SUPPLEMENTARY MATERIALS
Supplementary Material 1.
-
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare for this study.
-
FUNDING
This work was supported by Ministerio de Ciencia, Tecnología e innovación (MinCiencias Project: 111577756947).
-
AUTHOR CONTRIBUTIONS
Conceptualization: Montoya-Urrego D, Vanegas JM, Jiménez Quiceno JN. Formal analysis: Montoya-Urrego D, Vanegas JM, Jiménez-Quiceno JN. Funding acquisition: Jiménez Quiceno JN, Vanegas JM. Methodology: Montoya-Urrego D, Tellez-Carrasquilla S, Vanegas JM, Jiménez-Quiceno JN. Writing – original draft: Montoya-Urrego D, Tellez-Carrasquilla S. Writing – review & editing: Vanegas JM, Jiménez Quiceno JN.
NOTES
ACKNOWLEDGEMENTS

| Variable | Bivariate analysis | p-value | Multivariate analysis1 | p-value | |
|---|---|---|---|---|---|
| Market in the neighborhood store | 0.71 (0.33, 1.53) | 0.386 | 0.65 (0.31, 1.37) | 0.262 | |
| Preparation of food by the mother | 2.31 (1.52, 3.49) | <0.001 | 2.79 (1.30, 5.97) | 0.008 | |
| Travel last year | 0.94 (0.39, 2.26) | 0.886 | 0.89 (0.39, 1.99) | 0.769 | |
| Consumption of foods of animal origin | |||||
| Pork | 1.70 (0.52, 5.59) | 0.382 | 1.66 (0.49, 5.57) | 0.414 | |
| Beef | 1.07 (0.49, 2.35) | 0.863 | 1.04 (0.46, 2.33) | 0.925 | |
| Chicken | 0.83 (0.44, 1.59) | 0.579 | 0.79 (0.40, 1.56) | 0.499 | |
| Tap water consumption | 0.56 (0.31, 1.00) | 0.048 | 0.57 (0.32, 1.02) | 0.059 | |
| Handwashing | |||||
| Before eating | 0.91 (0.43, 1.90) | 0.792 | 1.01 (0.49, 2.08) | 0.988 | |
| After toilet | 0.50 (0.36, 0.70) | <0.001 | 0.46 (0.30, 0.71) | <0.001 | |
| Not washing hands | 1.94 (1.40, 2.70) | <0.001 | 1.89 (1.38, 2.58) | <0.001 | |
| History in last year | |||||
| Home medicine | 0.92 (0.47, 1.83) | 0.819 | 1.00 (0.48, 2.06) | 0.990 | |
| Hospitalization | 1.71 (0.87, 3.36) | 0.117 | 1.64 (0.83, 3.23) | 0.154 | |
| Surgery | 1.38 (0.72, 2.61) | 0.332 | 1.44 (0.77, 2.71) | 0.252 | |
| Antibiotic consumption | 2.17 (1.05, 4.47) | 0.036 | 2.13 (1.04, 4.36) | 0.038 | |
| Third-generation cephalosporins or fluoroquinolones | 1.16 (0.52, 2.60) | 0.714 | 1.10 (0.48, 2.50) | 0.816 | |
| Comorbidities: | |||||
| Cancer | 1.94 (1.40, 2.70) | <0.001 | 2.57 (1.42, 4.63) | 0.002 | |
| Diabetes mellitus | 1.80 (1.00, 3.23) | 0.048 | 2.13 (1.17, 3.85) | 0.013 | |
| Variables | Bivariate analysis | p-value | Multivariate analysis1 | p-value |
|---|---|---|---|---|
| Market in the neighborhood store | 4.79 (1.36, 16.83) | 0.015 | 4.54 (1.33, 15.53) | 0.016 |
| Preparation of food by the mother | 1.94 (0.62, 6.11) | 0.256 | 2.25 (0.73, 6.96) | 0.158 |
| Travel last year | 0.53 (0.14, 1.98) | 0.345 | 0.55 (0.15, 1.96) | 0.357 |
| Consumption of foods of animal origin | ||||
| Pork | 1.42 (0.35, 5.71) | 0.624 | 1.35 (0.33, 5.50) | 0.680 |
| Chicken | 0.42 (0.21, 0.82) | 0.011 | 0.43 (0.23, 0.82) | 0.011 |
| Tap water consumption | 0.83 (0.39, 1.77) | 0.627 | 0.80 (0.37, 1.72) | 0.570 |
| Handwashing | ||||
| Before eating | 0.74 (0.33, 1.66) | 0.467 | 0.76 (0.35, 1.65) | 0.494 |
| After toilet | 0.66 (0.15, 2.81) | 0.570 | 0.63 (0.14, 2.85) | 0.546 |
| History in the last year | ||||
| Home medicine | 0.95 (0.24, 3.72) | 0.945 | 1.00 (0.29, 3.50) | 0.994 |
| Hospitalization | 0.46 (0.08, 2.58) | 0.379 | 0.48 (0.13, 1.79) | 0.275 |
| Surgery | 0.43 (0.12, 1.50) | 0.185 | 0.48 (0.14, 1.60) | 0.232 |
| Antibiotic consumption | 0.86 (0.34, 2.19) | 0.752 | 0.93 (0.37, 2.33) | 0.874 |
| Comorbidities | ||||
| Diabetes mellitus | 1.87 (0.69, 5.11) | 0.221 | 2.68 (0.91, 7.89) | 0.073 |
| Sharing towels with the hemodialysis patient | 1.94 (0.94, 4.01) | 0.072 | 1.85 (0.91, 3.75) | 0.088 |
| Sharing soap with the hemodialysis patient | 1.28 (0.68, 2.39) | 0.441 | 1.36 (0.71, 2.59) | 0.352 |
| Sharing a bathroom with the hemodialysis patient | 1.89 (0.69, 5.15) | 0.214 | 1.87 (0.66, 5.25) | 0.237 |
| Sharing meals with the hemodialysis patient | 1.21 (0.87, 1.68) | 0.261 | 1.21 (0.88, 1.67) | 0.235 |
| Cleaning up the hemodialysis patient | 0.55 (0.24, 1.30) | 0.175 | 0.61 (0.26, 1.41) | 0.248 |
- 1. Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States; 2019 [cited 2022 Jan 11]. Available from: https://www.cdc.gov/drugresistance/biggest-threats.html.
- 2. Schwaber MJ, Carmeli Y. Mortality and delay in effective therapy associated with extended-spectrum beta-lactamase production in Enterobacteriaceae bacteraemia: a systematic review and metaanalysis. J Antimicrob Chemother 2007;60:913-920.PubMed
- 3. Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009;22:582-610.ArticlePubMedPMCPDF
- 4. Vanegas JM, Salazar-Ospina L, Montoya-Urrego D, Builes J, Roncancio GE, Jiménez JN. High frequency of colonization by diverse clones of beta-lactam-resistant Gram-negative bacilli in haemodialysis: different sources of transmission outside the renal unit? J Med Microbiol 2020;69:1132-1144.ArticlePubMed
- 5. Livermore DM. Current epidemiology and growing resistance of gram-negative pathogens. Korean J Intern Med 2012;27:128-142.ArticlePubMedPMC
- 6. Centers for Disease Control and Prevention. Recommendations for preventing transmission of infections among chronic hemodialysis patients; 2001 [cited 2022 Jan 11]. Available from: https://www.cdc.gov/mmwr/preview/mmwrhtml/rr5005a1.htm.Article
- 7. Calfee DP. Multidrug-resistant organisms in dialysis patients. Semin Dial 2013;26:447-456.ArticlePubMedPDF
- 8. Arcilla MS, van Hattem JM, Bootsma MC, van Genderen PJ, Goorhuis A, Schultsz C, et al. The Carriage of Multiresistant Bacteria after Travel (COMBAT) prospective cohort study: methodology and design. BMC Public Health 2014;14:410.ArticlePubMedPMCPDF
- 9. Viau R, Frank KM, Jacobs MR, Wilson B, Kaye K, Donskey CJ, et al. Intestinal carriage of carbapenemase-producing organisms: current status of surveillance methods. Clin Microbiol Rev 2016;29:1-27.ArticlePubMedPMCPDF
- 10. Centers for Disease Control and Prevention. Laboratory protocol for detection of carbapenem-resistant or carbapenemase-producing, Klebsiella spp. and E. coli from rectal swabs; 2008 [cited 2022 Jan 13]. Available from: https://www.aab.org/images/aab/pdf/2013/Klebsiella_or_Ecoli_Lab_protocol.pdf.
- 11. Clinical and Laboratory Standards Institute. M100: performance standards for antimicrobial susceptibility testing. 29th ed. Wayne: Clinical and Laboratory Standards Institute; 2019. p 32-42.
- 12. Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 2010;65:490-495.PubMed
- 13. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 2011;70:119-123.ArticlePubMed
- 14. Woodford N, Ellington MJ, Coelho JM, Turton JF, Ward ME, Brown S, et al. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Int J Antimicrob Agents 2006;27:351-353.ArticlePubMed
- 15. Durmaz R, Otlu B, Koksal F, Hosoglu S, Ozturk R, Ersoy Y, et al. The optimization of a rapid pulsed-field gel electrophoresis protocol for the typing of Acinetobacter baumannii, Escherichia coli and Klebsiella spp. Jpn J Infect Dis 2009;62:372-377.ArticlePubMed
- 16. Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 2006;60:1136-1151.ArticlePubMedPMC
- 17. Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep 2013;5:58-65.ArticlePubMed
- 18. Wendt R, Nickel O, Botsch A, Lindner M, Bethge A, Marx K, et al. Low colonization rates with Multidrug-resistant Gram-negative bacteria in a German hospital-affiliated hemodialysis center. PLoS One 2020;15:e0240314.ArticlePubMedPMC
- 19. Mota R, Pinto M, Palmeira J, Gonçalves D, Ferreira H. Multidrugresistant bacteria as intestinal colonizers and evolution of intestinal colonization in healthy university students in Portugal. Access Microbiol 2020;3:acmi000182.ArticlePubMedPMC
- 20. Maharjan A, Bhetwal A, Shakya S, Satyal D, Shah S, Joshi G, et al. Ugly bugs in healthy guts! Carriage of multidrug-resistant and ESBL-producing commensal Enterobacteriaceae in the intestine of healthy Nepalese adults. Infect Drug Resist 2018;11:547-554.ArticlePubMedPMCPDF
- 21. Bezabih YM, Sabiiti W, Alamneh E, Bezabih A, Peterson GM, Bezabhe WM, et al. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J Antimicrob Chemother 2021;76:22-29.ArticlePubMedPDF
- 22. Alconcher LF, Rivas M, Lucarelli LI, Galavotti J, Rizzo M. Shiga toxin-producing Escherichia coli in household members of children with hemolytic uremic syndrome. Eur J Clin Microbiol Infect Dis 2020;39:427-432.ArticlePubMedPDF
- 23. Adler A, Baraniak A, Izdebski R, Fiett J, Salvia A, Samso JV, et al. A multinational study of colonization with extended spectrum β-lactamase-producing Enterobacteriaceae in healthcare personnel and family members of carrier patients hospitalized in rehabilitation centres. Clin Microbiol Infect 2014;20:O516-O523.ArticlePubMed
- 24. Hernández-Gómez C, Blanco VM, Motoa G, Correa A, Vallejo M, Villegas MV, et al. Evolution of antimicrobial resistance in Gram negative bacilli from intensive care units in Colombia. Biomedica 2014;34 Suppl 1:91-100 (Spanish).PubMed
- 25. Mughini-Gras L, Dorado-García A, van Duijkeren E, van den Bunt G, Dierikx CM, Bonten MJ, et al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet Health 2019;3:e357-e369.ArticlePubMed
- 26. Karkaba A, Hill K, Benschop J, Pleydell E, Grinberg A. Carriage and population genetics of extended spectrum β-lactamaseproducing Escherichia coli in cats and dogs in New Zealand. Vet Microbiol 2019;233:61-67.ArticlePubMed
- 27. Toombs-Ruane LJ, Benschop J, French NP, Biggs PJ, Midwinter AC, Marshall JC, et al. Carriage of extended-spectrum-beta-lactamase- and AmpC beta-lactamase-producing Escherichia coli strains from humans and pets in the same households. Appl Environ Microbiol 2020;86:e01613-e01620.ArticlePubMedPMCPDF
- 28. Leimbach A, Hacker J, Dobrindt U. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 2013;358:3-32.ArticlePubMed
- 29. Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 2010;8:207-217.ArticlePubMedPDF
- 30. Dias RC, Vieira MA, Moro AC, Ribolli DF, Monteiro AC, Camargo CH, et al. Characterization of Escherichia coli obtained from patients undergoing peritoneal dialysis and diagnosed with peritonitis in a Brazilian centre. J Med Microbiol 2019;68:1330-1340.ArticlePubMed
- 31. Riley LW. Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin Microbiol Infect 2014;20:380-390.ArticlePubMed
- 32. Costa RF, Ferrari ML, Bringer MA, Darfeuille-Michaud A, Martins FS, Barnich N. Characterization of mucosa-associated Escherichia coli strains isolated from Crohn’s disease patients in Brazil. BMC Microbiol 2020;20:178.ArticlePubMedPMCPDF
- 33. Dogan B, Belcher-Timme HF, Dogan EI, Jiang ZD, DuPont HL, Snyder N, et al. Evaluation of Escherichia coli pathotypes associated with irritable bowel syndrome. FEMS Microbiol Lett 2018;365:fny249.Article
- 34. Muller A, Gbaguidi-Haore H, Cholley P, Hocquet D, Sauget M, Bertrand X. Hospital-diagnosed infections with Escherichia coli clonal group ST131 are mostly acquired in the community. Sci Rep 2021;11:5702.ArticlePubMedPMCPDF
- 35. Vega-Manriquez XD, Ubiarco-López A, Verdugo-Rodríguez A, Hernández-Chiñas U, Navarro-Ocaña A, Ahumada-Cota RE, et al. Pet dogs potential transmitters of pathogenic Escherichia coli with resistance to antimicrobials. Arch Microbiol 2020;202:1173-1179.ArticlePubMedPDF
- 36. Frost I, Van Boeckel TP, Pires J, Craig J, Laxminarayan R. Global geographic trends in antimicrobial resistance: the role of international travel. J Travel Med 2019;26:taz036.ArticlePubMedPDF
- 37. Bezabih YM, Sabiiti W, Alamneh E, Bezabih A, Peterson GM, Bezabhe WM, et al. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J Antimicrob Chemother 2021;76:22-29.ArticlePubMedPDF
- 38. Jiménez A, Alvarado A, Gómez F, Carrero G, Fajardo C. Risk factors associated with the isolation of extended spectrum betalactamases producing Escherichia coli or Klebsiella pneumoniae in a tertiary care hospital in Colombia. Biomedica 2014;34 Suppl 1:16-22 (Spanish).PubMed
- 39. Vanegas JM, Salazar-Ospina L, Roncancio GA, Builes J, Jiménez JN. Post-antibiotic era in hemodialysis? Two case reports of simultaneous colonization and bacteremia by multidrug-resistant bacteria. J Bras Nefrol 2021;43:597-602.ArticlePubMedPMC
- 40. Rada AM, Hernández-Gómez C, Restrepo E, Villegas MV. Distribution and molecular characterization of beta-lactamases in Gram-negative bacteria in Colombia, 2001-2016. Biomedica 2019;39(s1):199-220.ArticlePubMedPDF
- 41. Ghaddar N, Anastasiadis E, Halimeh R, Ghaddar A, Matar GM, Abou Fayad A, et al. Phenotypic and genotypic characterization of extended-spectrum beta-lactamases produced by Escherichia coli colonizing pregnant women. Infect Dis Obstet Gynecol 2020;2020:4190306.ArticlePubMedPMCPDF
- 42. Munasinghe T, Vidanapathirana G, Kuthubdeen S, Ekanayake A, Angulmaduwa S, De Silva K, et al. Colonization with selected antibiotic resistant bacteria among a cohort of Sri Lankan university students. BMC Infect Dis 2021;21:578.ArticlePubMedPMCPDF
- 43. Abdallah HM, Alnaiemi N, Reuland EA, Wintermans BB, Koek A, Abdelwahab AM, et al. Fecal carriage of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae in Egyptian patients with community-onset gastrointestinal complaints: a hospital-based cross-sectional study. Antimicrob Resist Infect Control 2017;6:62.ArticlePubMedPMCPDF
- 44. van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: an update. Infect Dis Clin North Am 2020;34:709-722.PubMedPMC
- 45. Salazar-Ospina L, Vanegas JM, Jiménez JN. High intermittent colonization by diverse clones of β-lactam-resistant Gram-negative bacilli suggests an excessive antibiotic use and different sources of transmission in haemodialysis patients. J Hosp Infect 2021;107:76-86.ArticlePubMed
- 46. Rezende TF, Doi AM, Quiles MG, Pignatari AC, Manfrendi S, Grothe C, et al. Detection of colonization by carbapenem-resistant organisms by real-time polymerase chain reaction from rectal swabs in patients with chronic renal disease. J Hosp Infect 2017;96:123-128.ArticlePubMed
REFERENCES
Figure & Data
References
Citations

- Molecular and Clinical Data of Antimicrobial Resistance in Microorganisms Producing Bacteremia in a Multicentric Cohort of Patients with Cancer in a Latin American Country
Sergio Andrés Cruz-Vargas, Laura García-Muñoz, Sonia Isabel Cuervo-Maldonado, Carlos Arturo Álvarez-Moreno, Carlos Humberto Saavedra-Trujillo, José Camilo Álvarez-Rodríguez, Angélica Arango-Gutiérrez, Julio César Gómez-Rincón, Katherine García-Guzman, Aur
Microorganisms.2023; 11(2): 359. CrossRef

 KSE
KSE
 PubReader
PubReader ePub Link
ePub Link Cite
Cite